Correlation plot matrices using the ellipse library
My new favorite library is the ellipse library. It includes functions for creating ellipses from various objects. It has a function, plotcorr() to create a correlation matrix where each correlation is represented with an ellipse approximating the shape of a bivariate normal distribution with the same correlation. While the function itself works well, I wanted a bit more redundancy in my plots and modified the code. I kept (most of) the main features provided by the function and I’ve included a few: the ability to plot ellipses and correlation values on the same plot, the ability to manipulate what is placed along the diagonal and the rounding behavior of the numbers plotted. Here is an example with some color manipulations. The colors represent the strength and direction of the correlation, -1 to 0 to 1, with University of Rochester approved red to white to blue.
First the function code:
my.plotcorr <- function (corr, outline = FALSE, col = "grey", upper.panel = c("ellipse", "number", "none"), lower.panel = c("ellipse", "number", "none"), diag = c("none", "ellipse", "number"), digits = 2, bty = "n", axes = FALSE, xlab = "", ylab = "", asp = 1, cex.lab = par("cex.lab"), cex = 0.75 * par("cex"), mar = 0.1 + c(2, 2, 4, 2), ...)
{
# this is a modified version of the plotcorr function from the ellipse package
# this prints numbers and ellipses on the same plot but upper.panel and lower.panel changes what is displayed
# diag now specifies what to put in the diagonal (numbers, ellipses, nothing)
# digits specifies the number of digits after the . to round to
# unlike the original, this function will always print x_i by x_i correlation rather than being able to drop it
# modified by Esteban Buz
if (!require('ellipse', quietly = TRUE, character = TRUE)) {
stop("Need the ellipse library")
}
savepar <- par(pty = "s", mar = mar)
on.exit(par(savepar))
if (is.null(corr))
return(invisible())
if ((!is.matrix(corr)) || (round(min(corr, na.rm = TRUE), 6) < -1) || (round(max(corr, na.rm = TRUE), 6) > 1))
stop("Need a correlation matrix")
plot.new()
par(new = TRUE)
rowdim <- dim(corr)[1]
coldim <- dim(corr)[2]
rowlabs <- dimnames(corr)[[1]]
collabs <- dimnames(corr)[[2]]
if (is.null(rowlabs))
rowlabs <- 1:rowdim
if (is.null(collabs))
collabs <- 1:coldim
rowlabs <- as.character(rowlabs)
collabs <- as.character(collabs)
col <- rep(col, length = length(corr))
dim(col) <- dim(corr)
upper.panel <- match.arg(upper.panel)
lower.panel <- match.arg(lower.panel)
diag <- match.arg(diag)
cols <- 1:coldim
rows <- 1:rowdim
maxdim <- max(length(rows), length(cols))
plt <- par("plt")
xlabwidth <- max(strwidth(rowlabs[rows], units = "figure", cex = cex.lab))/(plt[2] - plt[1])
xlabwidth <- xlabwidth * maxdim/(1 - xlabwidth)
ylabwidth <- max(strwidth(collabs[cols], units = "figure", cex = cex.lab))/(plt[4] - plt[3])
ylabwidth <- ylabwidth * maxdim/(1 - ylabwidth)
plot(c(-xlabwidth - 0.5, maxdim + 0.5), c(0.5, maxdim + 1 + ylabwidth), type = "n", bty = bty, axes = axes, xlab = "", ylab = "", asp = asp, cex.lab = cex.lab, ...)
text(rep(0, length(rows)), length(rows):1, labels = rowlabs[rows], adj = 1, cex = cex.lab)
text(cols, rep(length(rows) + 1, length(cols)), labels = collabs[cols], srt = 90, adj = 0, cex = cex.lab)
mtext(xlab, 1, 0)
mtext(ylab, 2, 0)
mat <- diag(c(1, 1))
plotcorrInternal <- function() {
if (i == j){ #diag behavior
if (diag == 'none'){
return()
} else if (diag == 'number'){
text(j + 0.3, length(rows) + 1 - i, round(corr[i, j], digits=digits), adj = 1, cex = cex)
} else if (diag == 'ellipse') {
mat[1, 2] <- corr[i, j]
mat[2, 1] <- mat[1, 2]
ell <- ellipse(mat, t = 0.43)
ell[, 1] <- ell[, 1] + j
ell[, 2] <- ell[, 2] + length(rows) + 1 - i
polygon(ell, col = col[i, j])
if (outline)
lines(ell)
}
} else if (i >= j){ #lower half of plot
if (lower.panel == 'ellipse') { #check if ellipses should go here
mat[1, 2] <- corr[i, j]
mat[2, 1] <- mat[1, 2]
ell <- ellipse(mat, t = 0.43)
ell[, 1] <- ell[, 1] + j
ell[, 2] <- ell[, 2] + length(rows) + 1 - i
polygon(ell, col = col[i, j])
if (outline)
lines(ell)
} else if (lower.panel == 'number') { #check if ellipses should go here
text(j + 0.3, length(rows) + 1 - i, round(corr[i, j], digits=digits), adj = 1, cex = cex)
} else {
return()
}
} else { #upper half of plot
if (upper.panel == 'ellipse') { #check if ellipses should go here
mat[1, 2] <- corr[i, j]
mat[2, 1] <- mat[1, 2]
ell <- ellipse(mat, t = 0.43)
ell[, 1] <- ell[, 1] + j
ell[, 2] <- ell[, 2] + length(rows) + 1 - i
polygon(ell, col = col[i, j])
if (outline)
lines(ell)
} else if (upper.panel == 'number') { #check if ellipses should go here
text(j + 0.3, length(rows) + 1 - i, round(corr[i, j], digits=digits), adj = 1, cex = cex)
} else {
return()
}
}
}
for (i in 1:dim(corr)[1]) {
for (j in 1:dim(corr)[2]) {
plotcorrInternal()
}
}
invisible()
}
And now a short walk through:
#usage of my.plotcorr
#much like the my.plotcorr function, this is modified from the plotcorr documentation
#this function requires the ellipse library, though, once installed you don't need to load it - it is loaded in the function
#install.packages(c('ellipse'))
#library(ellipse)
source('my.plotcorr.R')
# Get some data
data(mtcars)
# Get the correlation matrix
corr.mtcars <- cor(mtcars)
# Change the column and row names for clarity
colnames(corr.mtcars) = c('Miles/gallon', 'Number of cylinders', 'Displacement', 'Horsepower', 'Rear axle ratio', 'Weight', '1/4 mile time', 'V/S', 'Transmission type', 'Number of gears', 'Number of carburetors')
rownames(corr.mtcars) = colnames(corr.mtcars)
# Standard plot, all ellipses are grey, nothing is put in the diagonal
my.plotcorr(corr.mtcars)
# Here we play around with the colors, colors are selected from a list with colors recycled
# Thus to map correlations to colors we need to make a list of suitable colors
# To start, pick the end (and mid) points of a scale, here a red to white to blue for neg to none to pos correlation
colsc=c(rgb(241, 54, 23, maxColorValue=255), 'white', rgb(0, 61, 104, maxColorValue=255))
# Build a ramp function to interpolate along the scale, I've opted for the Lab interpolation rather than the default rgb, check the documentation about the differences
colramp = colorRampPalette(colsc, space='Lab')
# I'll show two types of color styles using this color ramp
# the first
# Use the same number of colors along the scale for the number of variables
colors = colramp(length(corr.mtcars[1,]))
# then plot an example with only ellipses, without a diagonal and with a main title
# the color selection stuff here multiplies the correlations such that they can index individual colors and create a sufficiently large list
# incase you are confused, r allows vector indexing with non-integers by rounding down, i.e. colors[1.8] == colors[1]
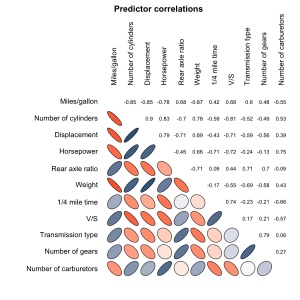
my.plotcorr(corr.mtcars, col=colors[5*corr.mtcars + 6], main='Predictor correlations')
# the second form
# we could, alternatively, make a scale with 100 points
colors = colramp(100)
# then pick colors along this 100 point scale given the correlation value * 100 rounded down to the nearest integer
# to do that we need to move the correlation range from [-1, 1] to [0, 100]
# now plot again with ellipses along the diagonal
my.plotcorr(corr.mtcars, col=colors[((corr.mtcars + 1)/2) * 100], diag='ellipse', main='Predictor correlations')
# or, add numbers to the bottom of the chart
my.plotcorr(corr.mtcars, col=colors[((corr.mtcars + 1)/2) * 100], diag='ellipse', lower.panel="number", main='Predictor correlations')
# or, switch the numbers and ellipses and reduce the margins
my.plotcorr(corr.mtcars, col=colors[((corr.mtcars + 1)/2) * 100], diag='ellipse', upper.panel="number", mar=c(0,2,0,0), main='Predictor correlations')
# or, drop the diagonal and numbers
my.plotcorr(corr.mtcars, col=colors[((corr.mtcars + 1)/2) * 100], upper.panel="none", mar=c(0,2,0,0), main='Predictor correlations')
March 26, 2012 at 7:29 pm
Nice!
LikeLike
April 11, 2013 at 6:25 am
awesome! perfect visualization 🙂
LikeLike
May 22, 2013 at 8:26 am
Very nice plot,
but I tried to put my own data set, the R showed
> data(XXX)
Warning message:
In data(XXX) : data set ‘XXX’ not found
I am pretty sure I have imported my dataset in.
Would you mind helping me to sort it out?
LikeLike
May 22, 2013 at 2:22 pm
Hi Roger,
the only call to data is “data(mtcars)”. mtcars is a data set that comes with R. I just tried that command in R 3.0 and it works. So, I am not sure what causes the problem. Or did you type “data(XXX)”? That would not work, even if you have created a data set called XXX. The command “data()” loads an data set that comes with a library in R. if you have already loaded your own data set. Just remove that command from the code.
hth,
Florian
LikeLike
May 23, 2013 at 4:25 am
Sorry, probably let you feel confused,
load(XXX) , XXX means my own data set’ name, I know mtcars is built in R, so I changed the name,
I mean I’d like to import my own data and make a beautiful plot as yours,
but it showed:
> data(XXX)
Warning message:
In data(XXX) : data set ‘XXX’ not found
so I was just wondering that did I miss anything?
Cheers,
LikeLike
May 23, 2013 at 10:10 am
Hi Roger,
As Florian mentioned, the data() function is just to load an example data set that comes with an R library—not your own data. I am not sure what ‘XXX’ stands for in your question. If you have a saved dataset (in an .RData or similar R native file) you’ll need to use the load() function on that file. If your data is in a raw text format you should load it using the read.table() function. As an example, if you have your data in a tab delimited file named ‘MyData.tab’ you can replace the data() line with something like this:
my.data = read.table(file="path/to/MyData.tab", sep="\t") #be sure to check for row and column name issues
#and continue in a similar way through the rest of the code.
corr.my.data = cor(my.data)
#be sure to change any variable names and other specifics for your data in the rest of the code
-Esteban
LikeLike
July 22, 2013 at 2:05 am
Thanks for the nice code!
LikeLike
November 7, 2013 at 4:49 am
The numerical values are not displayed when i run the code, any help?
Thanks
lemma
LikeLike
November 7, 2013 at 11:19 am
Hi Lemma,
I’m not sure what could be the issue. The numbers should be displayed if you specify that you want numbers in the lower left or upper right half of the plot. For example, using the mtcars data above you can put numbers in the upper right half like this:
my.plotcorr(corr.mtcars, upper.panel="number")LikeLike
December 3, 2013 at 12:13 pm
This is a very nice modification of the plotcorr function, thanks! Might I suggest two things: 1) Perhaps include an option to plot histograms for single variables along the diagonal, and 2) options for significance stars on correlation values. Just suggestions…
LikeLike
December 3, 2013 at 5:51 pm
Thanks for the suggestions. Do you have any specific code in mind that you’d be willing to share?
LikeLike
January 29, 2014 at 6:45 am
Hello Everyone.
Sorry, if I post this question here, but I’m very new to R.
How can I make this function run in my program? Do I have to kind of install it, or just insert the function code and everything into my scriptfile and press the button?
Do I have to install this function somewhere in the library of ellipses?
Thanks for your help (maybe a link would be helpfull )
)
LikeLike
January 29, 2014 at 4:48 pm
You just paste the function into your script window, read it in (or ‘source’ it) and then you should be able to call it. You can also set up R so that it sources a specific script file every time it starts.
HTH
LikeLike
January 30, 2014 at 4:21 am
Well, that is what I’ve tried so far.
I took the whole code (from function code, not just the one line where the function is defined) and paste it into my script window and read it in.
But then, when it comes to the “application”, when I tipe in source(‘my.plotcorr.R’), it says: Cannot open Connection. The ellipse-library is installed of course.
Ty for help 🙂
LikeLike
January 30, 2014 at 8:30 am
I’m not quite sure what you’re doing but I’d suggest that you copy paste the function code from the post into a file called ‘my.plotcorr.R’ and save it somewhere. Then in a separate script file where you want to generate plots, source that file (i.e. with source(‘my.plotcorr.R’)). Make sure you also give the source function the right path to that file otherwise R will give you the error “cannot open connection”. Alternatively set R’s working directory to the same one as where that file is. I keep all my helper scripts in the same place on my computer so it’s easy for source whatever I need from any other script on my computer.
LikeLike
March 31, 2014 at 8:42 pm
Hi, I’ve been using the great modified function for a while and everything was working until today.
I’m getting the following error: “Error in ellipse(mat, t = 0.43) : center must be a vector of length 2”
When I use the original ellipse package, it’s fine, but the modified version is giving me the error. Any tips?
Thanks,YB
LikeLike
April 24, 2014 at 3:57 pm
Hi JB- looks like we got the same error. It’s because of another package with a different ellipse function (probably car, as it is in my case)
LikeLike
June 30, 2014 at 8:47 am
I’m getting the following error message too: “Error in ellipse(mat, t = 0.43) : center must be a vector of length 2″
I tried to solve the masking problem by adding ellipse : : ellipse() befor the function but I still receive the error. Any ideas? Thank you!
LikeLike
June 30, 2014 at 9:37 am
Could you give me a minimal (non-) working example to see if I can reproduce the issue?
LikeLike
August 1, 2014 at 2:51 pm
Hello, First of thanks for the original post!
I’m trying to apply this to a dataset that has some missing values under a few of the variable. In the correlation matrix the r value shows up as NA for any correlations including Variable that have any missing values.
I done see a natural place to insert something like “na.rm = TRUE” to have it carry out the correlation analysis even if values are missing.
Thanks in advance!
LikeLike
August 1, 2014 at 3:22 pm
It seems this function needs a correlation matrix as input, so you’d have to put in the na.rm in the call that creates the correlation matrix (what did you use)?
LikeLike
August 2, 2014 at 11:06 pm
Thanks for the comment Florian. The plotting function does not calculate correlations for you so how you calculate them is left to the user but cor() doesn’t have an na.rm argument.
TLab, you’ll need to add the use=’complete’ option for your cor(). This will find complete cases to calculate your correlation matrix. In the case of the walkthrough above it’d be on line 10 like this:
corr.mcars <- cor(mtcars, use='complete')
Alternatively pass your data.matrix through the function complete.cases() before you pass it to cor() will also work. Just be aware that these solutions will remove rows for which there is any missing data so if you want to keep as much data as possible for each pairwise correlation you'll need to calculate each on their own and build the square correlation matrix to pass to the plotting function.
HTH,
Esteban
LikeLike
October 5, 2014 at 10:22 am
thanks a lot for the post! worked like wonders. is there a way to get the size of the correlation coefficients appear bigger on the chart? thanks!
LikeLike
October 5, 2014 at 4:06 pm
This is something that isn’t quite intuitive to do given the code as is, the best you can do is to reduce the size of the labels and any text in the plot (with the cex.lab and cex parameters). Doing this will scale the size of the ellipses relative to everything else.
HTH!
LikeLike
March 28, 2017 at 10:19 am
I am dealing with the same issue – where exactly to I need to use the cex/cex.lab option to scale up the size of the text? I’ve tried setting cex = 2 instead of cex.lab in row 45.46, but that didn’t seem to do anything…
LikeLike
April 14, 2015 at 12:48 pm
Hello! Thank you for sharing this code. Should it be cited/referenced in any specific way if I use it in a publication?
LikeLike
April 14, 2015 at 5:32 pm
Feel free to cite the Core R team and Ellipses library. You can use the
citation()command in R to see what version is appropriate.LikeLike
April 14, 2015 at 7:32 pm
And pointers to this post or blog ate always appreciated 🙂
LikeLike