Plotting effects for glmer(, family=”binomial”) models
UPDATE 12/15/10: Bug fix. Thanks to Christian Pietsch.
UPDATE 10/31/10: Some further updates and bug fixes. The code below is the updated one.
UPDATE 05/20/10: I’ve updated the code with a couple of extensions (both linear and binomial models should now work; the plot now uses ggplot2) and minor fixes (the code didn’t work if the model only had one fixed effect predictor). I also wanted to be clear that the dashed lines in the plots aren’t confidence intervals. They are multiples of the standard error of the effect.
Here’s a new function for plotting the effect of predictors in multilevel logit models fitted in R using lmer() from the lme4 package. It’s based on code by Austin Frank and I also borrowed from Harald Baayen’s plotLMER.fnc() (package languageR). First a cool pic:
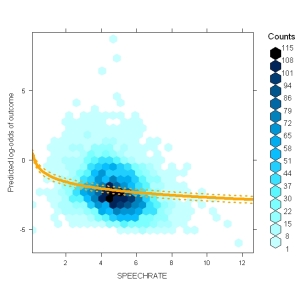
These plots contain the distribution of the predictor (x-axis) against the predicted values (based on the entire model, y-axis) using hexbinplot() from the package hexbin. On top of that, you see the model prediction fo the selected predictor along with confidence intervals. Note that the predictor is given in its original form (here speech rate) although it was entered into the model as the centered log-transformed speechrate. The plot consideres that. Of course, you can configure things.
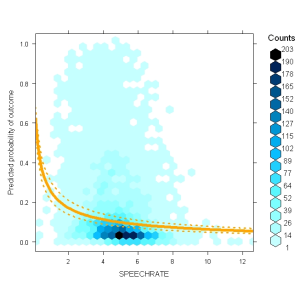
For example, you could plot the effect in probability space:
my.glmergplot <- function(
# version 0.43
# written by tiflo@csli.stanford.edu
# code contributions from Austin Frank, Ting Qian, and Harald Baayen
# remaining errors are mine (tiflo@csli.stanford.edu)
#
# last modified 12/15/10
#
# now also supports linear models
# backtransforms centering and standardization
#
# known bugs:
# too simple treatment of random effects
#
model,
name.predictor,
name.outcome= "outcome",
predictor= NULL,
# is the predictor centered IN THE MODEL?
# is the predictor transformed before
# (centered and) ENTERED INTO THE MODEL?
predictor.centered= if(!is.null(predictor)) { T } else { F },
predictor.standardized= F,
predictor.transform= NULL,
fun= NULL,
type= "hex",
main= NA,
xlab= NA,
ylab= NA,
xlim= NA,
ylim= NA,
legend.position="right",
fontsize=16,
col.line= "#333333",
col.ci= col.line,
lwd.line= 1.2,
lty.line= "solid",
alpha.ci= 3/10,
hex.mincnt= 1,
hex.maxcnt= nrow(model@frame) / 2,
hex.limits = c(round(hex.mincnt), round(hex.maxcnt)),
hex.limits2 = c(round(match.fun(hex.trans)(hex.mincnt)), round(match.fun(hex.trans)(hex.maxcnt))),
hex.midpoint = (max(hex.limits) - (min(hex.limits) - 1)) / 2,
hex.nbreaks = min(5, round(match.fun(hex.trans)(max(hex.limits)) - match.fun(hex.trans)(min(hex.limits))) + 1),
hex.breaks = round(seq(min(hex.limits), max(hex.limits), length.out=hex.nbreaks)),
hex.trans = "log10",
...
)
{
if (!is(model, "mer")) {
stop("argument should be a mer model object")
}
if ((length(grep("^glmer", as.character(model@call))) == 1) &
(length(grep("binomial", as.character(model@call))) == 1)) {
model.type = "binomial"
} else {
if (length(grep("^lmer", as.character(model@call))) == 1) {
model.type = "gaussian"
}
}
if (!(model.type %in% c("binomial","gaussian"))) {
stop("argument should be a glmer binomial or gaussian model object")
}
if (!is.na(name.outcome)) {
if (!is.character(name.outcome))
stop("name.outcome should be a string\n")
}
if (!is.na(xlab[1])) {
if (!is.character(xlab))
stop("xlab should be a string\n")
}
if (!is.na(ylab)) {
if (!is.character(ylab))
stop("ylab should be a string\n")
}
# load libaries
require(lme4)
require(Design)
require(ggplot2)
if (predictor.standardized) { predictor.centered = T }
if (is.null(fun)) {
if (is.na(ylab)) {
if (model.type == "binomial") { ylab= paste("Predicted log-odds of", name.outcome) }
if (model.type == "gaussian") { ylab= paste("Predicted ", name.outcome) }
}
fun= I
} else {
if (is.na(ylab)) {
if (model.type == "binomial") { ylab= paste("Predicted probability of", name.outcome) }
if (model.type == "gaussian") { ylab= paste("Predicted ", name.outcome) }
}
fun= match.fun(fun)
}
if (!is.null(predictor.transform)) {
predictor.transform= match.fun(predictor.transform)
} else { predictor.transform= I }
indexOfPredictor= which(names(model@fixef) == name.predictor)
# get predictor
if (is.null(predictor)) {
# simply use values from model matrix X
predictor= model@X[,indexOfPredictor]
# function for predictor transform
fun.predictor= I
if (is.na(xlab)) { xlab= name.predictor }
} else {
# make sure that only defined cases are included
predictor = predictor[-na.action(model@frame)]
# function for predictor transform
trans.pred = predictor.transform(predictor)
m= mean(trans.pred, na.rm=T)
rms = sqrt(var(trans.pred, na.rm=T) / (sum(ifelse(is.na(trans.pred),0,1)) - 1))
fun.predictor <- function(x) {
x= predictor.transform(x)
if (predictor.centered == T) { x= x - m }
if (predictor.standardized == T) { x= x / rms }
return(x)
}
if ((is.na(xlab)) & (label(predictor) != "")) {
xlab= label(predictor)
}
}
# get outcome for binomial or gaussian model
if (model.type == "binomial") {
outcome= fun(qlogis(fitted(model)))
} else {
outcome= fun(fitted(model))
}
## calculate grand average but exclude effect to be modeled
## (otherwise it will be added in twice!)
## random effects are all included, even those for predictor (if any).
## should random slope terms for the predictor be excluded?
## prediction from fixed effects
if (ncol(model@X) > 2) {
Xbeta.hat = model@X[, -indexOfPredictor] %*% model@fixef[-indexOfPredictor]
} else {
Xbeta.hat = model@X[, -indexOfPredictor] %*% t(model@fixef[-indexOfPredictor])
}
## adjustment from random effects
Zb = crossprod(model@Zt, model@ranef)@x
## predicted value using fixed and random effects
Y.hat = Xbeta.hat + Zb
## intercept is grand mean of predicted values
## (excluding fixed effect of predictor)
## (including random effects of predictor, if any)
int = mean(Y.hat)
# slope
slope <- fixef(model)[name.predictor]
## error and confidence intervals
stderr <- sqrt(diag(vcov(model)))
names(stderr) <- names(fixef(model))
slope.se <- stderr[name.predictor]
lower <- -1.96 * slope.se
upper <- 1.96 * slope.se
# setting graphical parameters
if (is.na(ylim)) { ylim= c(min(outcome) - 0.05 * (max(outcome) - min(outcome)), max(outcome) + 0.05 * (max(outcome) - min(outcome)) ) }
if (is.na(xlim)) { xlim= c(min(predictor) - 0.05 * (max(predictor) - min(predictor)), max(predictor) + - 0.05 * (max(predictor) - min(predictor))) }
print("Printing with ...")
print(paste(" int=", int))
print(paste(" slope=", slope))
print(paste(" centered=", predictor.centered))
print(" fun:")
print(fun.predictor)
pdata= data.frame( predictor=predictor, outcome=outcome )
x= seq(xlim[1], xlim[2], length=1000)
fit= int + slope * fun.predictor(x)
ldata= data.frame(
predictor= x,
outcome= fun(fit),
transformed.lower= fun(fit + lower),
transformed.upper= fun(fit + upper)
)
theme_set(theme_grey(base_size=fontsize))
theme_update(axis.title.y=theme_text(angle=90, face="bold", size=fontsize, hjust=.5, vjust=.5))
theme_update(axis.title.x=theme_text(angle=0, face="bold", size=fontsize, hjust=.5, vjust=.5))
p <- ggplot(data=pdata, aes(x=predictor, y=outcome)) +
xlab(xlab) +
ylab(ylab) +
xlim(xlim) +
ylim(ylim) +
opts(legend.position=legend.position, aspect.ratio=1)
# for degbugging:
# panel.lines(rep(mean(x),2), c(min(y),max(y)))
# panel.lines(c(min(x),max(x)), c(mean(y),mean(y)))
if (type == "points") {
p <- p + geom_point(alpha=3/10)
} else if (type == "hex") {
p <- p + geom_hex(bins = 30) +
scale_fill_gradient2(low= "lightyellow",
mid="orange",
high=muted("red"),
midpoint= hex.midpoint,
space="rgb",
name= "Count",
limits= hex.limits,
breaks= hex.breaks,
trans = hex.trans
)
}
p + geom_ribbon(data=ldata,
aes( x= predictor,
ymin=transformed.lower,
ymax=transformed.upper
),
fill= col.ci,
alpha= alpha.ci
) +
geom_line(data=ldata,
aes(x= predictor,
y=outcome
),
colour= col.line,
size= lwd.line,
linetype= lty.line,
alpha=1
)
}
The only thing this function really needs is a model and a NAME of a predictor, e.g. this plots the distribution of centered log-transformed speechrate against the predicted probability of a complementizer (the outcome of my model). On top of the distribution, it plots the prediction dependent on the value of cLSPEECHRATE and the CIs around it. Per default this plots in log-odds space.
I think this is a great way plot to detect outliers – everything is in the right space: the y-axis is the average predicted log-odds (=fitted(model)) and the x-axis is in the scale actually used in the model
my.glmerplot(lmer.speaker, "cLSPEECHRATE") # but you can go to probability space (now debugged) my.glmerplot(lmer.speaker, "cLSPEECHRATE", fun=plogis)
The above plots are nice since they plot against the actual scale of the predictor, but they are often hard to interpret.
What if you want the nice scale from your original variable on the x-axis? that is, if you don’t want centered log-transformed speechrate, but rather speechrate? The distribution would then be nicely plotted in the space you can understand (but beware: to detect relevant outliers, see above). The predicted effect should also be plotted correctly — so, we need to tell the model what was done to the original input variable and we need to give it the original variable:
my.glmerplot(lmer.speaker, "cLSPEECHRATE", predictor= d$SPEECHRATE, predictor.transform = log, predictor.centered=T) # or shorter (predictor.centered defaults to T) my.glmerplot(lmer.speaker, "cLSPEECHRATE", predictor= d$SPEECHRATE, predictor.transform = log) # or in probability space my.glmerplot(lmer.speaker, "cLSPEECHRATE", predictor= d$SPEECHRATE, predictor.transform = log, fun= plogis)
The following, however, would be wrong (since variable IS actually centered and logged in model). Usually that becomes apparent when you plot, but be cautious
my.glmerplot(lmer.speaker, "cLSPEECHRATE", predictor= d$SPEECHRATE, predictor.centered=F) my.glmerplot(lmer.speaker, "cLSPEECHRATE", predictor= d$SPEECHRATE)
of course, you can modify all the color settings and other parameters. see above. Here is an example:
my.glmerplot(lmer.speaker, "cLSPEECHRATE", fun=plogis,
xlab="Centered log-transformed speechrate (syllables/second)",
name.outcome="complementizer",
col.line = "black",
col.int = "gray",
colramp = function (n) { plinrain(n, beg=20, end=225) })
Which, in all ugliness, would look like:
I am sure there are plenty of bugs in there or room for improvement and extension. Please comment on this post, or send us your updated improved version =).
Hhhmm, probably it would be better to include an option to plot the “real” distribution, i.e. not against the predicted (fitted) values, but against true averages in the “bin”. If you know what I mean …
November 18, 2009 at 4:50 pm
Hi,
I used your new code to plot standard error lines from the logistic regression by lmer. However, it seems that predicted lines (with standard error lines) depart significantly from predicted values. If needed, I can send my picture to you, so you can point out where my mistake is.
Thank you very much!
Best wishes,
Jianghua
LikeLike
November 18, 2009 at 7:06 pm
Hi Jianghua, yes please send me your plot and the code you used. That would be very helpful! (fjaeger@bcs.rochester.edu). Thanks.
Florian
LikeLike
June 14, 2010 at 10:31 pm
Can my.glmergplot plot interaction effects as well? Also, this function seems to only take predictors that are continuous. What if I have a predictor that has been sum-coded?
LikeLike
June 14, 2010 at 10:47 pm
Hi Alan,
right now, this does not do interactions (I have some other code for that). Since I always code my categorical predictors per hand anyway (to center them), I hadn’t thought of handling factors, but I should. That should be easy enough. As soon as I’ll be done with grant writing I should add this. If you guys come up with something before then, please let me know and we can post it here. That would be of interest to lots of folks, I bet.
florian
LikeLike
October 31, 2010 at 12:11 pm
Any updates to this great function with interactions? I’m trying to plot varying slopes nicely …
LikeLike
October 31, 2010 at 2:29 pm
Hi bshor,
I think Neal Snider has some code for including interactions. I’ll point him to this post and maybe he can upload it. I also have made some updates to the function that I will upload to the above page in a sec.
florian
LikeLike
November 1, 2010 at 10:53 am
Great! By the way, the formatting is a little screwed up on the code here (beginning with line creating the function fun.predictor), so copying and pasting leads to errors.
LikeLike
November 1, 2010 at 3:56 pm
thanks for noticing! hopefully it’s fixed now.
LikeLike
December 15, 2010 at 5:36 am
Hi Florian, thank you for sharing this code.
I am trying to use it (version 0.42) for plotting a GLMM with logit link. However, it produces the following error message and aborts:
So the source of the problem seems to be this line of code:
As far as I can tell,
predictorstill has its default value (NULL) at this point. Or is it an obligatory argument? How is this supposed to work?For completeness, here is some more information on my model:
In the end, I was able to produce a plot after commenting out the line of code quoted above. Perhaps it should be re-inserted further down?
Cheers,
Christian
LikeLike
December 16, 2010 at 1:01 am
Hi Christian –
thanks for catching this. You’re right, the statement should be applied later. It’s necessary when you supply a value for the (non-obligatory) predictor parameter. By specifying the predictor parameter you can plot variables on the original scale. E.g. if you logged and centered something but want interpretable x-axis values, you provide the predictor parameter, predictor.centered=T, predictor.transform = log.
The line that caused the problem (now moved downwards in the code; see above) makes sure that only those rows of the predictor with defined values are included (since those are the only ones that are included in the model’s frame).
Florian
LikeLike
January 3, 2011 at 9:48 am
[…] Plotting effects for glmer(, family=”binomial”) models January 2009 10 comments […]
LikeLike
January 31, 2011 at 4:02 pm
Hi Florian, thanks for the code…it’s working quite nicely for me.
However, I do have one question. Is there a simple way for me to adapt the
code so that the hexbinplot() code plots the raw values that were fed into
the LMER model, while still maintaining the line of fit and confidence
intervals from the predictor values?
I would it would have something with changing the following lines, but I’m
not sure:
pdata= data.frame( predictor=predictor, outcome=outcome )
p <- ggplot(data=pdata, aes(x=predictor, y=outcome)) +
xlab(xlab) +
ylab(ylab) +
xlim(xlim) +
ylim(ylim) +
opts(legend.position=legend.position, aspect.ratio=1)
Thanks,
Kyle Lovseth
LikeLike
January 31, 2011 at 4:55 pm
Hi Kyle,
I am not sure that I understand your question. The function can plot raw values for the predictor (x-axis) at least for some transformations. Say, you have a log-transformed predictor, then you would do what is shown at the top of the post (you’ll give the predictor on the original scale, along with a transformation function; cf. the first plot above which keeps the original scale of speech rate, but plots a log-transformed effect).
Or did you mean, having the outcome on the original scale? That wouldn’t really work well without additional assumptions for binomial models.
Florian
LikeLike
January 31, 2011 at 5:33 pm
Yes, sorry. I should have specified that I wanted raw values for the Outcome. I’m rather new to posting within technical blogs;)
My model is linear, and I was looking for a way to illustrate how well the predicted slope of my model fits the raw data, without having a monstrous “cloud” of data points on the graph.
Anyways, I think I figured it out by adding the following changes to your script:
raw = model@y
pdata= data.frame( predictor=predictor, outcome=raw )
I have VERY little programming experience, but I think this should work, no?
Thanks again,
Kyle Lovseth
LikeLike
January 31, 2011 at 7:26 pm
That should work. Some folks in my lab have adapted the function for linear models. I’ll try to get some of them to comment.
Florian
LikeLike
February 23, 2011 at 7:45 pm
Hi,
I tried to use the function earlier, but got an error message as below:
Error in my.glmergplot(answer.glmer, “ms”) :
could not find function “fun”
The model I fitted is below:
answer.glmer <- glmer(answer~ms*es + (1|subject)+(1|item), alldata, subset = p == 11, family = "binomial")
I wonder what might've gone wrong.
Thanks!
Xiao
LikeLike
February 23, 2011 at 9:07 pm
Dear Xiao, does the function work for other models?
Florian
LikeLike
November 7, 2011 at 5:11 pm
Hi,
i run the function to plot the partial effects of a model that i did with the package lme4. But it gives me an error: “Error in model@X[, -indexOfPredictor] %*% t(model@fixef[-indexOfPredictor]) :
no compatible arguments”
I will apreciatte if you can tell what is the problem. Thanks.
Rafael
LikeLike
November 7, 2011 at 9:46 pm
Hi Rafael,
I need a little bit more detail, e.g. an example of the data and the code you applied to it (incl. the model and the call to the function). As a guess, could it be that your model only had one fixed effect predictor in it?
Florian
LikeLike
March 1, 2012 at 9:09 am
Hi Florian,
I went back to work with the function “my.glmergplot”. I have a model, where my dependent variable is binomial. I have only one independent variable (categorical with 2 levels), and two random intercepts. When i run the function it gives me the same error that I presented in the previous post:
“Error in model@X[, -indexOfPredictor] %*% t(model@fixef[-indexOfPredictor]) :
no compatible arguments”
I used the lmer function, and my model run without problems.
Thanks in advance, kind regards,
Rafael.
LikeLike
March 4, 2012 at 3:46 pm
Hi Francisco,
unfortunately, I am leaving town for conferences. At this point, I’d really need to know more about your data and script. Perhaps you could email me (to tiflo@csli.stanford.edu) a subset of your data (with only the relevant variables as coded in the model) and the model call as well as the call to the plotting function? Then I can try to help (though not before 4/1, I think ==)).
Florian
LikeLike
June 13, 2012 at 6:57 pm
Hi – I noticed that the Design package is not available these days, and has been replaced by rms package. Can rms simply be replaced into the function? Also, I find I get the error,
Error in -na.action(model@frame) : invalid argument to unary operator. I am pretty sure I am naming my predictor properly. Is there any common reason for this error?
Thanks,
Arthur
LikeLike
June 16, 2012 at 3:16 pm
Hey Arthur,
sorry, I was offline for a few days. yes, Design is now rms. Many function still work the same way but some things have been re-organized. I am not sure whether this would affect this function. Did you get the error running an old version of R with the Design package or a new version with the rms library?
florian
LikeLike
July 3, 2012 at 12:42 pm
Hi folks,
Really hope you can help me on this. As I am not a statistician and I am fairly new to R programming language I wonder if you could provide some pointers. I have searched high and low for ways to plot my mixed effect glmer model and found very little until now. The model deals with the presence and absence of an invasive species on navigation buoys (pa_cm). The fixed effect variables are the percentage cover of different animal/plants on the buoys (f1….f4). As there was more than one sample taken per buoy I want to include this as a random effect (Num). In addition buoys came from different geographical areas with possible different conditions so I want to include Area as a random effect.
So this is the model I am using
fita<-glmer(pa_cm~f1+f2+f3+f4+(1|Num)+(1|Area),family=binomial("logit"),data=data)
The key question is how do I include my model output in your plotting script, Forgive my ignorance, but how do I state my model and predictor in this case. If you have any input on the model or how best to plot it I would be over the moon. Best wishes.
Adrain
LikeLike
July 3, 2012 at 4:50 pm
Try
fita<-glmer(pa_cm~f1+f2+f3+f4+(1|Num)+(1|Area),family=binomial("logit"),data=data)
my.glmerplot(fita, "f1", predictor.centered=F, predictor.standardized=F)
or, if you did center your predictors,
my.glmerplot(fita, "f1", predictor.centered=T, predictor.standardized=F)
Let me know whether these work.
Florian
LikeLike
July 4, 2012 at 7:26 am
Hey Floran,
Thanks for getting back to me. For both I get a Error: could not find function “my.glmerplot” . I have look through the code to make sure the scrip is running OK. It seems to all go through without any warning messages. One possible reason could be that in the latest version of R the ‘Design’ package is unavailable. Best wishes.
Adrian
LikeLike
July 4, 2012 at 1:34 pm
Yes, the function is terribly outdated. If you look at the comment right below yours, you’ll see that Ted had the same problem and changed the function to work with the new Design package (now called “rms”). He offered to post the updated code here and once he has done that, I’ll look through it and update the page (with credits to him, of course). It shouldn’t take many changes though. So, stay tuned!
LikeLike
July 3, 2012 at 5:01 pm
Hi Florian
seems I changed (or maybe “kludged” is a better word) your script and replaced the Design package with ez. Works great for R 2.14 . Seems R 2.15 has killed the script. I have a laptop still running 2.14 so I am OK. I can send you the script and data set if you are interested.
LikeLike
July 3, 2012 at 5:10 pm
Hi Ted,
yes, pls post it here. Thanks!
Florian
LikeLike
July 23, 2012 at 7:03 am
great function! thanks, Florian!
LikeLike
February 19, 2015 at 3:17 pm
This looks extremely useful. I don’t know how much has changed in the past two years, but I was just trying to get this function to work and I’m having some trouble. It looks like model$fixef is old syntax for fixef(model) and same for ranef, but I don’t know what model@X is for, nor what model@x or model@Zt is for. Could you please explain those bits or let me know if you think this will be too difficult to update?
Thank you!
LikeLike
February 19, 2015 at 5:37 pm
Dear Emily,
model@X is/was the data matrix (the matrix that binds all input predictors as columns, and cases as rows). model@Zt is the transpose of the random-effects model matrix. Apparently, this structure has changed since lme4.0 (which the above function was made for). The help page says:
More generally, you can find all of these components by googling for “lmer” and the component’s name (e.g., “Zt”).
hth,
Florian
LikeLike
May 28, 2015 at 3:45 am
Dear Florian and Emily,
It seems that I was able to adapt all the code to moder packages/slots/stuff, but one line. I just can’t figure out how to adapt the following:
Zb = crossprod(model@Zt, model@ranef)@x
I mean, I already know that the first part can be somewhat achieved by:
as.matrix(getME(model, “Zt”))
While the other part, in case of only one hierarchical level, can be achieved by:
as.matrix(ranef(model)[[1]])
However, the cross-product between these two pieces is impossible. Besides, there is no slot X anymore in any of these matrices, so I don’t know exactly where it should be. Any ideas? We are almost there. 🙂
Thanks in advance for your time.
LikeLike
May 28, 2015 at 4:20 am
I think I got even closer, but there is still something missing: the color gradient does not appear and the counting of cases seems off. Here I send my code so far (feel free to cut it from the comment when moderating, since it is going to get too big a comment):
my.glmergplot <- function(
# version 0.43
# written by tiflo@csli.stanford.edu
# code contributions from Austin Frank, Ting Qian, and Harald Baayen
# remaining errors are mine (tiflo@csli.stanford.edu)
#
# last modified 12/15/10
#
# now also supports linear models
# backtransforms centering and standardization
#
# known bugs:
# too simple treatment of random effects
#
model,
name.predictor,
name.outcome= "outcome",
predictor= NULL,
# is the predictor centered IN THE MODEL?
# is the predictor transformed before
# (centered and) ENTERED INTO THE MODEL?
predictor.centered= if(!is.null(predictor)) { T } else { F },
predictor.standardized= F,
predictor.transform= NULL,
fun= NULL,
type= "hex",
main= NA,
xlab= NA,
ylab= NA,
xlim= NA,
ylim= NA,
legend.position="right",
fontsize=16,
col.line= "#333333",
col.ci= col.line,
lwd.line= 1.2,
lty.line= "solid",
alpha.ci= 3/10,
hex.mincnt= 1,
hex.maxcnt= nrow(model@frame) / 2,
hex.limits = c(round(hex.mincnt), round(hex.maxcnt)),
hex.limits2 = c(round(match.fun(hex.trans)(hex.mincnt)), round(match.fun(hex.trans)(hex.maxcnt))),
hex.midpoint = (max(hex.limits) – (min(hex.limits) – 1)) / 2,
hex.nbreaks = min(5, round(match.fun(hex.trans)(max(hex.limits)) – match.fun(hex.trans)(min(hex.limits))) + 1),
hex.breaks = round(seq(min(hex.limits), max(hex.limits), length.out=hex.nbreaks)),
hex.trans = "log10",
…
)
{
#if (!is(model, "mer")) {
# stop("argument should be a mer model object")
#}
if ((length(grep("^glmer", as.character(model@call))) == 1) &
(length(grep("binomial", as.character(model@call))) == 1)) {
model.type = "binomial"
} else {
if (length(grep("^lmer", as.character(model@call))) == 1) {
model.type = "gaussian"
}
}
if (!(model.type %in% c("binomial","gaussian"))) {
stop("argument should be a glmer binomial or gaussian model object")
}
if (!is.na(name.outcome)) {
if (!is.character(name.outcome))
stop("name.outcome should be a string\n")
}
if (!is.na(xlab[1])) {
if (!is.character(xlab))
stop("xlab should be a string\n")
}
if (!is.na(ylab)) {
if (!is.character(ylab))
stop("ylab should be a string\n")
}
# load libaries
require(lme4)
require(rms)
require(ggplot2)
require(scales)
if (predictor.standardized) { predictor.centered = T }
if (is.null(fun)) {
if (is.na(ylab)) {
if (model.type == "binomial") { ylab= paste("Predicted log-odds of", name.outcome) }
if (model.type == "gaussian") { ylab= paste("Predicted ", name.outcome) }
}
fun= I
} else {
if (is.na(ylab)) {
if (model.type == "binomial") { ylab= paste("Predicted probability of", name.outcome) }
if (model.type == "gaussian") { ylab= paste("Predicted ", name.outcome) }
}
fun= match.fun(fun)
}
if (!is.null(predictor.transform)) {
predictor.transform= match.fun(predictor.transform)
} else { predictor.transform= I }
indexOfPredictor= which(names(fixef(model)) == name.predictor)
# get predictor
if (is.null(predictor)) {
# simply use values from model matrix X
predictor= getME(model, "X")[,indexOfPredictor]
# function for predictor transform
fun.predictor= I
if (is.na(xlab)) { xlab= name.predictor }
} else {
# make sure that only defined cases are included
predictor = predictor[-na.action(model@frame)]
# function for predictor transform
trans.pred = predictor.transform(predictor)
m= mean(trans.pred, na.rm=T)
rms = sqrt(var(trans.pred, na.rm=T) / (sum(ifelse(is.na(trans.pred),0,1)) – 1))
fun.predictor 2) {
Xbeta.hat = getME(model, “X”)[, -indexOfPredictor] %*% fixef(model)[-indexOfPredictor]
} else {
Xbeta.hat = getME(model, “X”)[, -indexOfPredictor] %*% t(fixef(model)[-indexOfPredictor])
}
## adjustment from random effects
Zb = crossprod(as.matrix(getME(model, “Zt”)), unlist(ranef(model)[[1]])) #@x
## predicted value using fixed and random effects
Y.hat = Xbeta.hat + Zb
## intercept is grand mean of predicted values
## (excluding fixed effect of predictor)
## (including random effects of predictor, if any)
int = mean(Y.hat)
# slope
slope <- fixef(model)[name.predictor]
## error and confidence intervals
stderr <- sqrt(diag(vcov(model)))
names(stderr) <- names(fixef(model))
slope.se <- stderr[name.predictor]
lower <- -1.96 * slope.se
upper <- 1.96 * slope.se
# setting graphical parameters
if (is.na(ylim)) { ylim= c(min(outcome) – 0.05 * (max(outcome) – min(outcome)), max(outcome) + 0.05 * (max(outcome) – min(outcome)) ) }
if (is.na(xlim)) { xlim= c(min(predictor) – 0.05 * (max(predictor) – min(predictor)), max(predictor) + – 0.05 * (max(predictor) – min(predictor))) }
print("Printing with …")
print(paste(" int=", int))
print(paste(" slope=", slope))
print(paste(" centered=", predictor.centered))
print(" fun:")
print(fun.predictor)
pdata= data.frame( predictor=predictor, outcome=outcome )
x= seq(xlim[1], xlim[2], length=1000)
fit= int + slope * fun.predictor(x)
ldata= data.frame(
predictor= x,
outcome= fun(fit),
transformed.lower= fun(fit + lower),
transformed.upper= fun(fit + upper)
)
theme_set(theme_grey(base_size=fontsize))
theme_update(axis.title.y=element_text(angle=90, face="bold", size=fontsize, hjust=.5, vjust=.5))
theme_update(axis.title.x=element_text(angle=0, face="bold", size=fontsize, hjust=.5, vjust=.5))
p <- ggplot(data=pdata, aes(x=predictor, y=outcome)) +
xlab(xlab) +
ylab(ylab) +
xlim(xlim) +
ylim(ylim) +
theme(legend.position=legend.position, aspect.ratio=1)
# for degbugging:
# panel.lines(rep(mean(x),2), c(min(y),max(y)))
# panel.lines(c(min(x),max(x)), c(mean(y),mean(y)))
if (type == "points") {
p <- p + geom_point(alpha=3/10)
} else if (type == "hex") {
p <- p + geom_hex(bins = 30) +
scale_fill_gradient2(low= "lightyellow",
mid="orange",
high=muted("red"),
midpoint= hex.midpoint,
space="rgb",
name= "Count",
limits= hex.limits,
breaks= hex.breaks,
trans = hex.trans
)
}
p + geom_ribbon(data=ldata,
aes( x= predictor,
ymin=transformed.lower,
ymax=transformed.upper
),
fill= col.ci,
alpha= alpha.ci
) +
geom_line(data=ldata,
aes(x= predictor,
y=outcome
),
colour= col.line,
size= lwd.line,
linetype= lty.line,
alpha=1
)
}
LikeLike
November 15, 2015 at 5:31 am
I think that people should be aware that this function requires models that are mer objects not merMod objects which models are in the latest version of lme4
LikeLike
January 31, 2016 at 11:01 pm
I really like these plots, thanks for posting the code! – I’m receiving the following error message when I run the code…
argument should be a mer model object
(but my model is a mer object since it’s glmer
I run your code first, plus I load library(hexbin)
then, I run my code below:
y <- with(data1, cbind(Mean_Red, (Total – Mean_Red)))
prev <- with(data1, as.numeric(prevalence))
model.glmer <- glmer(y ~ prev + (1|lot) + (1|obs), family = binomial, data = data1)
my.glmerplot(model.glmer, "prev", predictor.centered = F, predictor.standardized = F, data = data1)
Please pardon my ignorance!
Many thanks in advance
LikeLike